Open Access Journal of Agriculture Research
(ISSN 2651-9003)
Research Article
Genome-Wide Identification of miRNAs Targets Involved in Cold Response in Cassava
Li S1, Peng M1* and Cheng Z2
1Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
2Haikou Experimental Station, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
*Corresponding author: Ming Peng, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou, China, Tel: +86089866890981; Email: pengming@itbb.org.cn
Citation: Li S1, Peng M, Cheng Z (2019) Genome-Wide Identification of miRNAs Targets Involved in Cold Response in Cassava. Open Acc J Agri Res: OAJAR-100020.
Received date: 9 October 2019; Accepted date: 16 October, 2019; Published date: 23 October, 2019
Abstract
MicroRNAs (miRNAs) are recognized as essential transcriptional or post-transcriptional regulators, and play versatile roles in plants, functioning in processes such as growth, development and stress responses. Cassava (Manihot esculenta) is a major root crop widely grown as a staple food, animal feed, and also as an important source of bioethanol worldwide. Cold stress seriously affects cassava plants growth, development and yield. MiRNAs and their targets have been extensively studied in model plants, but a genome-wide identification of miRNAs’ targets is still lacking in cassava. In this study, to identify the roles of miRNAs and their targets in response to cold stress, two degradome libraries were constructed using cold-treated and non-cold-treated cassava seedlings.
Degradome data allowed us to identify a total of 151 non-redundant miRNA-target pairs from the degradome data. We reveal that approximately 42% of miRNA targets are conserved across plant species. However, 83 novel miRNA targets were identified in the two libraries, suggesting a specific role for these genes in response to cold stress. Gene ontology analyses showed that many target genes involved in “cellular process” and “metabolic process”. In addition, 12 miRNAs and 31 corresponding targets of them were further found to be involved in cold stress response. Particularly, miR159, miR164 and miR396 participated in cold stress response by up-regulating certain MYBs, NACs and GRFs genes that were involved in the regulation of downstream gene expression. Meantime, three miRNAs and five target genes were validated by quantitative real-time PCR (qRT-PCR). The work helps identify cold-responsive miRNA targets in cassava and increases the number of novel targets involved in cold stress response. Furthermore, the findings of this study might provide valuable reference and new insights for understanding the functions of miRNA in stress response in plants.
Keywords: Cassava; Degradome Sequencing; miRNA; Target genes; qRT-PCR
Abbreviations
miRNAs : microRNAs
GO : Gene Ontology
qRT-PCR : Quantitative reverse transcription polymerase chain reaction.
TFs : Transcription Factors
Introduction
Cassava (Manihot esculenta) is a major root crop widely grown as a staple food, animal feed, and also as an important source of bioethanol in the tropical and subtropical regions of Latin America, Africa and Asia. It displays a unique ability to produce acceptable yields on marginal soils with minimal inputs and tolerate drought conditions. Nevertheless, as a tropical plant, cassava is categorized as a cold-sensitive species. Cold stress has been recognized as the most important limiting factor for its geographical location and productivity. Low temperatures and frozen conditions generally cause serious damage to cassava plants, such as reduced leaf expansion, chlorosis in leaves and yield decrease. In addition, the physiological status of cold-stressed cassava plants is also altered, such as transient increases in soluble sugars and proline levels, changes in membrane lipid composition, and increases in the level of reactive oxygen species (ROS) [1-5].
Thus, to stabilize cassava yield under unfavorable temperature stresses, it is necessary to identify key genes or pathways that can be used to improve cold stress tolerance of cassava via genetic engineering. So far, the mechanisms of cold stress response have been preliminarily investigated in the cassava. A multitude of putative cold stress tolerance-related genes have been identified and characterized by a combination of genetic, biochemical and high-throughput sequencing approaches. For examples, transgenic cassava plants that overexpress an ArabidopsisC-repeat-binding factors (CBFs) gene have shown enhanced tolerance to multiple abiotic stresses. Overexpression of two ROS-scavenging enzymes, cytosolic superoxide dismutase (SOD) and ascorbate peroxidase (APX), led to increased tolerance to oxidative and chilling stresses in cassava. These studies advanced our knowledge regarding the molecular genetic mechanisms underlying the cold stress response in cassava [6-10].
MicroRNAs (miRNAs) are a class of 20-24 nucleotide (nt) endogenous noncoding RNAs that play essential roles in regulating gene expression in animals, plants, and fungi. Plant miRNAs have been shown to play regulatory roles at the post-transcriptional level by directly degrading target messenger RNAs (mRNAs) or repressing translation [11]. In plants, miRNAs control the expression of genes encoding transcription factors, enzymes, and other important proteins, participate in the regulation of a wide range of biological processes such as normal growth, development, hormone signaling, nutrient homeostasis, and responses to various abiotic and biotic stresses. In the past decade, a large number of miRNAs and targets have been identified across several plant species [12]. For instance, miR156 regulates expression of MYB genes involved in the control of many aspects of plant development, such as flowering under short days, primary root growth and seed germination [13-15]. Moreover, miR164 targets several different NAC (NAM/ATAF/CUC) genes to regulate the formation of shoot meristems, age-dependent leaf senescence and the specification of organ boundaries. In the last decade, accumulating evidences place miRNAs and their targets in a central position within gene expression programs that underlie plant development and stress responses. Numerous studies have shown that plant miRNAs are involved in the regulation of cold response in plants, such as Arabidopsis, rice, maize, barley, wheat, cotton, and barrel medic. For examples, eighteen cold-responsive miRNAs were identified in rice, such as miRNA166, 169 and 319. Overexpression of miR319 or genetically down-regulating the expression of either of the two miR319-targeted genes, OsPCF5 and OsPCF8, leads to enhanced cold tolerance in rice [16-20].
Given the important role of miRNAs in plant stress response, we speculated that miRNA and their targets might remarkably contribute to the cold stress tolerance of cassava [21-25]. To date, approximately 48 miRNAs associated with cold stress were identified by establishing small RNA libraries of cassava. However, a genome-wide identification of miRNAs’ targets is still lacking. Further, we aimed to determine which miRNAs’ targets and corresponding pathways responded to cold stress to facilitate survival of cassava. To address this question, we performed high-throughput sequencing and quantitative reverse transcription polymerase chain reaction (qRT-PCR) to identify the conserved and cold-responsive miRNAs’ targets in cassava leaves [26-30]. A total of 151 miRNA-target pairs were identified using degradome sequencing and bioinformatics. Several conserved miRNAs’ targets and the responses of these genes to cold stress were further discussed. Among them, 3 cold-responsive miRNA and corresponding targets were identified by qRT-PCR analyses. The results of this study provide new insights for understanding the miRNA-mediated regulatory network associated with cold stress in cassava [31-35].
Results
Construction of the degradome library and sequencing Analysis
In order to identify the miRNA targets in cassava plants at a global level, two degradome libraries (Cold and Control library) which captured the cleaved mRNAs were constructed for sequencing by a Hiseq2000 platform [36-40]. A total of 24,256,967 and 24,655,669 raw tags were obtained from Control and cold samples, respectively. After filtering the low-quality tags, the total number of clean tags in each library ranged from 24,256,875 to 24,655,584 tags with a mean value of 99.9%. More than 98.5 % of the clean reads were successfully mapped to the cassava reference genome to identify the fragments of degraded mRNAs (Table 1). Moreover, approximately 34% clean reads were uniquely mapped to the genome. These data indicated that our two degradome libraries were of high quality and recovered most of the degraded mRNA targets that contained the sequence profile of miRNA-mediated cleavage and allowed us to conduct further analysis [41-45].
Systematic identification of miRNA targets: In plants, the majority of miRNA-guided post-transcriptional regulation cleaves the target mRNAs between the 10th and 11thnot from the 5’ of miRNA in the complementary region of the miRNA: mRNA pair. In cassava, conserved miRNA targets were previously investigated mainly by bioinformatics prediction and only a few miRNA targets have been experimentally validated. To better understand the functional roles of miRNAs, high-throughput degradome sequencing was performed in this study to identify more miRNA targets in cassava, particularly specific targets of cold-responsive miRNA targets [46-50].
A total of 33,028 known transcripts and 176 miRNA sequences of cassava were used to annotate the degradome tags, and 144 target genes of 20 conserved miRNA families were verified (Table 2). Based on the relative abundances of target site reads compared with other sites in the gene model, the miRNA targets were categorized into five classes (categories 0, 1, 2, 3 and 4). Under control condition, there were 83, 5, 32, 2 and 0 targets in categories 0, 1, 2, 3 and 4 respectively. Under cold condition, 88, 8, 20, 4 and 36 targets were grouped into categories 0, 1, 2, 3 and 4 respectively (Supplementary Table S1). In total, the majority of targets were grouped into categories of greater than the median confidence (categories 3 and 4) in our degradome sequencing, accounting for 75%. Finally, a total of 151 miRNA-target pairs, related to 20 miRNAs and 144 genes, were identified with a high confidence level (p value < 0.05) (Supplementary Table S1). Among the 20 identified conserved miRNA families, MIR156, MIR 166, MIR 172 and MIR396 targeted multiple genes, whereas no targets were discovered for 16 miRNA families, such as MIR390, MIR397, MIR828, MIR1711, MIR2111, MIR1446 and MIR3627. The identified targets of the conserved miRNAs included many previously identified targets, such as the TCP genes for miR319, SQUAMOSA promoter binding protein-like (SPL) genes as the targets of miR156, auxin response factor (ARF) as the target of miR167, and growth-regulating factors (GRFs) as the target of miR396 [51-55].
However, we also identified 83 novel target genes of miRNAs, including a lysine-specific histone demethylase (Manes.11G098200) for miR156, GTPase-activating protein for miR159, and a hypothetical protein (Manes.04G138500) as the target of miR396(Supplementary Table S2). Many of the target genes of the conserved miRNAs were classified as the transcription factors (TFs), such as SPLs, AP2s, NACs, ARFs, MYBs, HD-ZIPs and GRFs, and so on. These TFs are known to regulate diverse aspects of plant growth, development, and biotic and abiotic stresses. Compared with model plants, such as Arabidopsis and rice, many conserved miRNA targets were found to be conserved in cassava, indicating that the miRNA-target relationship was evolutionarily conserved [56-60].
GO analysis of miRNA target genes: To classify the functions of the identified target genes for miRNAs, we performed Gene Ontology (GO) functional classification analysis. Based to GO analysis, a total of 144 miRNA targets were classified into three categories: biological process (BP), cellular component (CC) and molecular function (MF), which were classified into 15, 13 and 9 terms respectively, as shown in Figure 1. Among these groups, the most significantly enriched GO terms were involved in the three main categories, “cellular process”, “cell” and “binding”, followed by “metabolic process”, “cell part” and “catalytic binding”. The results suggested that most target genes were associated with gene expression regulation and metabolic pathways.
Identification of miRNA targets in response to cold stress: In previous studies, we reported transcriptome changes in cassava in response to cold and identified over 6000 cold-responsive genes by analyzing the ssRNA-Seq data. To further investigate the expression pattern of miRNAs-targeted genes under cold stress, we compared the expression values of the identified targets of control samples and those of cold-treated samples in the ssRNA-Seq data. According to the criteria described previously, 31 miRNAs targets were determined as the differentially expressed genes in cassava under cold stress (Table 3). Among them, 23 miRNAs targets were identified to be up-regulated and 8 were down-regulated by cold treatment. As shown in Table 3, five miR164-targeted NAC family genes, including Manes.06G090500 and Manes.14G080400, were strongly up-regulated by cold stress.
This result is consistent with the expression pattern of miR164 which expression level was sharply declined to the lowest level at 24 h after cold treatment. MiR396 targeted a number of genes annotated as GRFs and NBS-LRR resistance proteins, which was also induced by cold stress. Altogether, these results indicated that the miR164, miR396 and family and their target genes played a key important function in responding to cold stress in cassava [61].
Detection of the expression of miRNA and their targets by qRT-PCR: To further reveal the dynamic expression pattern of miRNAs and their target genes, the expression profiles of 5 miRNA-mRNA pairs (miR159-MYB, miR164-NACs and miR396-GRFs) were selected, and qRT-PCR analyses were conducted to validate the alterations in their expression levels under cold conditions revealed by the high-throughput sequencing results. As shown in Figure 2, the expression levels of miR159 and miR396 decreased to the lowest value after cold treatment.
However, within the same time period, their corresponding targets MYB and GRFs exhibited apparently the opposite expression profile. The expression correlation of these miRNAs and their targets demonstrated that they negatively regulated their targets. We also found that the expression level of miR164 and NACs genes were surprisingly induced by cold treatment. Further studies will be needed to elucidate the molecular function of these miRNAs and their targets in response to cold stress in cassava.
Discussion
MiRNAs play essential roles in response to abiotic stress, especially in cold stress response. High throughput sequencing and computational approaches have been used in recent years for identifying a huge number of miRNAs and targets in a range of plant species. In cassava, however, miRNA targets were previously investigated mainly via bioinformatics prediction, and only a few conserved miRNA targets have been experimentally validated. Therefore, it is necessary to globally identify miRNA targets for interpretation of their functions. In the study, we constructed and sequenced two distinct degradome libraries using cassava leaves under normal and cold stress conditions. By analyzing degradome-Seq data, a large number of miRNA targets were successfully identified. These target transcripts generated significant GO terms related to binding and catalytic activities. Importantly, our degradome data verified 90% of previously validated targets. Additionally, our analysis revealed that the majority of these target genes were conserved among species, and were obviously enriched in TFs and transcription regulatory activity, such as SPLs, MYBs, HD-ZIPs, ARFs, GRFs and TCP. This phenomenon was similar to that found in other plant species.
Several targets of conserved miRNAs in this study, such as the SPL gene (target of miR156) and TCP gene (target of miR319), have been reported that are required for abiotic stresses responses in some other plant species, indicating the miRNAs and its targets may have a similar role in different plant species. In a previous study, eight conserved miRNAs (miR156, miR159, miR160, miR162, miR166, miR171, miR395, and miR396) were down-regulated under cold stress, while miR157, miR1168, and miR398 were up-regulated, and the corresponding targets were identified in our degradome sequencing. The gene encoding the MYB domain transcription factor is a conserved target of the miR159 family in plants. In Arabidopsis, miR159 have been verified to target encoding MYBs to regulate plant growth and abiotic stresses responses. In addition, MYB33 and MYB101 are targeted by miR159 and appear to play an important role in the response to abscisic acid (ABA) during seedling stress responses. In this study, we detected a number of MYB family TFs regulated by mes-miR159. MiR159 expression is down-regulated after cold treatment, and regulates the abundance of MYB mRNAs in response to cold stress. MiR164 is a conserved miRNA in many plant species and guide the cleavage of the mRNAs of five NAC transcription factor genes in Arabidopsis that are involved in lateral root emergence, and age-dependent cell death.Several studies have shown that miR164 is also involved in response to abiotic and biotic stress in plants.MiR164 showed slightly up-regulated expression in response to cold stress in this study, which was consistent with previous research. The miR396 and Growth-regulating factor (GRF) regulatory network is evolutionarily conserved among plant species. The GRFs is a plant specific family of transcription factors defined by the presence of the WRC and QLQ protein domains. Seven out of the 9 Arabidopsis GRFs have a binding site for microRNA (miRNA) miR396.GRFs have been implicated in the development and growth of plant organs and structures. Previous studies have revealed that miR396 is responsive to a variety of environmental stresses, such as salt, drought, oxidative stress and cold stress. Transgenic rice and Arabidopsis plants constitutively over-expressing osa-MIR396c showed reduced salt stress tolerance compared to that of wild-type plants. Our findings indicate that the expression of miR396 decreased significantly and its target genes was up-regulated during cold stress. To date, investigations on cassava miR396 and their targets are rather rare. Clarifying the biological functions of the miR396 targets in response to abiotic stresses will help uncover the function of miR396 in crops.
Materials and methods
Plant material and stress treatment: Cassava (Manihotesculenta) cultivar (60444) was used in the study. As previously described, the stems about 1.5 cm in size with one bud were cut and planted in MS plate for 2 weeks in a growth chamber at 26 ± 2?, with a photoperiod of 16 h light and 8 h dark. For cold treatment, plants with a uniform growth status were treated at 4? for 24 h in a chamber under 16 h light and 8 h dark.
The youngest leaves and shoot apex were collected from the same position of the plants and then were immediately frozen in liquid nitrogen for RNA extraction.This protocol was chosen for two reasons: (i) we measure the expression of cold stress marker genes (known DREB/CBFs and CORs/RD29) using RNA derived from samples exposed to cold stress at 4? for different time points, the expression of the marker genes is maximal at 24 hours after cold stress (Li et al., 2017b). (ii) we observed visible leave wilting in cold stress treatments (Supplementary Figure S1).Untreated plants at the same stage grown in chamber under 16 h light and 8 h dark were used as controls.
Library construction of degradome-seq and bioinformatic analysis: Two degradome libraries (Control and Cold), one replicate for each library, were constructed as described previously with minor modifications. The total RNA extraction, libraries construction and deep sequencing were performed by the Guangzhou Genedenovo Biotechnology Co., Ltd (Guangzhou, China).
First, poly (A) + mRNA was enriched by Oligo (dT) magnetic beads. Then, the cleavage products with a free 5’-monophosphate at their 3’ termini were ligated with 5’ RNA adapters, followed with reverse transcription and PCR. Two libraries were sequenced on an Illumina Hiseq 2000 platform. The raw reads were pre-processed to filter low-quality reads, adaptors, tRNAs, rRNAs, snRNAs and snoRNAs. To identify genes targeted by miRNAs, the remaining clean reads were mapped to the reference genes (cDNA) of cassava by SOAP2 tools, and the miRNA-mRNA pairs were searched and p-values were calculated using PAREsnip. Additionally, the CleaveLand program was run to detect potentially cleaved miRNA targets. All alignments with p-value < 0.05 that possessed the 5’ end of the degradome sequence coincident with the 10th and 11th nucleotides were considered as miRNA targets [62].
Functional annotations of the miRNA targets: To investigate the putative biological functions of target genes and biological processes possibly regulated by miRNAs in cassava, Gene Ontology (GO) analysis were employed to annotate and classify target genes. The GO categorization results were listed as three independent hierarchies for biological process, cellular component, and molecular function [63].
Detection of miRNA and their targets using qRT-PCR: Total RNA from the same samples was prepared using the RNA Plant kit (OMEGA). For miRNA, approximately 3μg of total RNA was reverse-transcribed using miRNA-specific stem-loop primers in a 20μL of reaction volume using a Fermentas RevertAid First trand cDNA Synthesis kit (Fermentas, USA). For miRNAs’ targets, the cDNA was generated using 2 μg of total RNA and OligodT18 primer with the PrimeScriptTM RT reagent kit (Takara). Real time qPCR assays were performed using the SYBR Premix ExTaqTM (Takara) followed the manuals from the manufacturers. U6 small nuclear RNA and MeACTIN gene were used as the internal control for miRNA and corresponding targets, respectively [64].
The expression level of the miRNAs and their targets in different samples were calculated by comparative 2-??CT method. Standard deviations were calculated from three biological replicates. The primers used for qRT-PCR analysis are listed in Supplementary Table 2.
Data access
The degradome RNA sequencing raw data has been deposited in the NCBI Sequence Read Archive (SRA) under the BioProject accession number SRP133534.
Conclusion
This is the first comprehensive identification of cold-responsive miRNAs targets in cassava by degradome sequencing. A total of 151 miRNA-target pairs were identified, including many new target miRNA pairs. This study provides important data for better understanding the molecular mechanisms of resistance to cold in cassava. Further experiments on the identified target genes are necessary to explore potential mechanisms of abiotic stress regulation in cassava.
Acknowledgments
We thank Dr. Xueyi Tian for helpful comments on this manuscript. This work was supported by the National Natural Science Foundation of China (31561143012, 31701484, and 31701903), the National Key R&D Program of China (2018YFD1000500), and the grants from the Young Elite Scientists Sponsorship Program by CSTC (CSTC-QN201702).
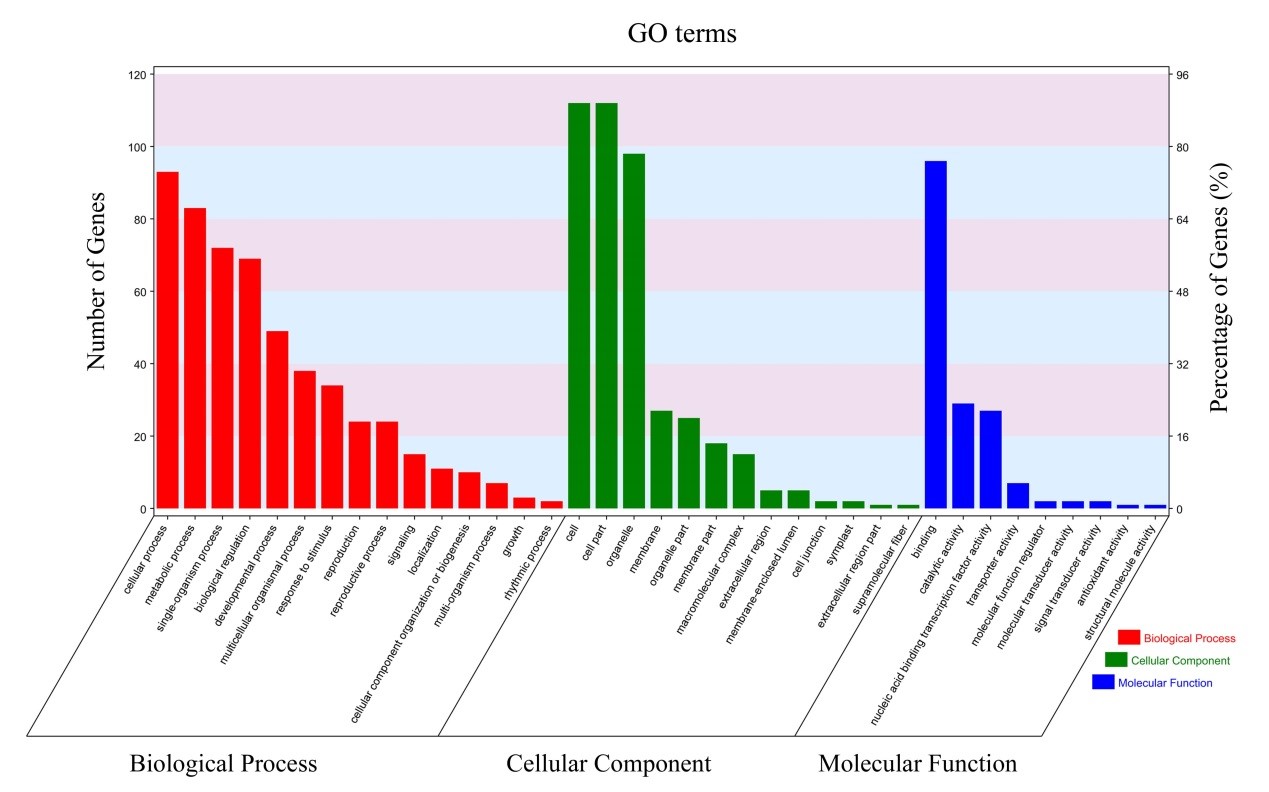
Figure 1: Gene Ontology functional classification of the miRNA targets identified in cassava. The x-axis represents the diverse biological functions of the targets according to three GO categories (biological process, cellular component and molecular function). The y-axis represents the number of the target genes.
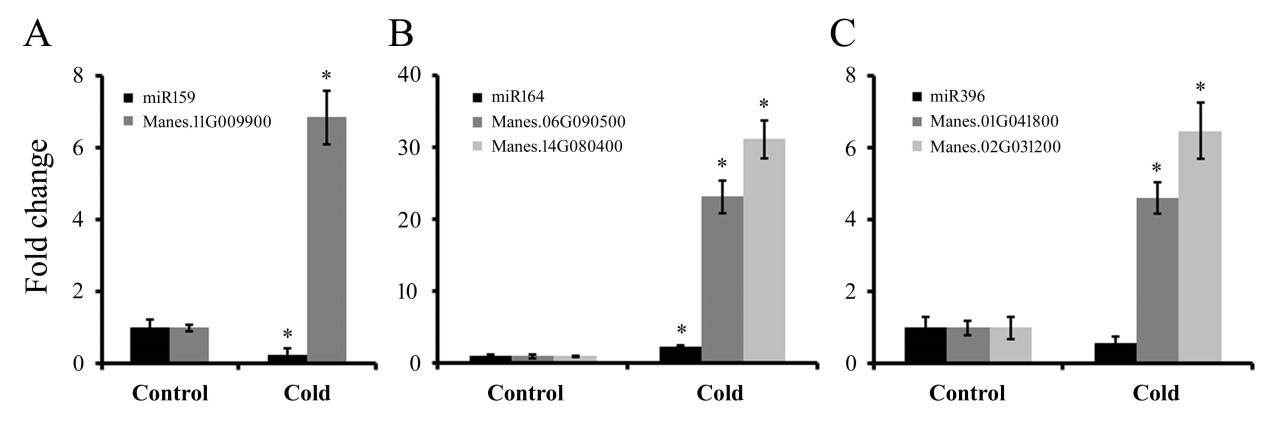
Figure 2: The expression patterns of three miRNAs and their targets under cold stress in cassava. U6 and MeACTIN were used as an internal control for miRNA and targets, respectively. Error bars represent ±SD from three independent experiments.
|
Type |
Control |
Cold |
|
Raw tags |
24,256,967 |
24,655,669 |
|
Clean tags |
24,256,875(99.9%) |
24,655,584(99.9%) |
|
Total mapped tags |
23,792,501(98.1%) |
24,291,679(98.5%) |
|
Unique mappable tags |
8,002,069(33.6%) |
8,595,344(35.3%) |
|
cDNA mapped tags |
11,420,405(48.9%) |
10,202,506(42.6%) |
Table 1: Data set summary of sequencing of the two degradome libraries.
|
miRNA |
Total Targets |
Conserved targets |
Targets ID |
|
miR156 |
36 |
7 SPLs |
Manes.01G026000 Manes.05G050800 Manes.09G032800 Manes.12G009000 Manes.12G010200 Manes.13G009400 Manes.13G011000 |
|
miR159 |
7 |
5 MYBs |
Manes.01G175900 Manes.02G133900 Manes.04G153700 Manes.05G052300 Manes.11G009900 |
|
miR160 |
9 |
7 ARFs |
Manes.04G019800 Manes.07G099400 Manes.07G099500 Manes.08G007600 Manes.10G046700 Manes.11G146500 Manes.18G072400 |
|
miR164 |
6 |
5 NACs |
Manes.02G047800 Manes.03G105700 Manes.06G090500 Manes.14G080400 Manes.15G089700 |
|
miR166 |
20 |
7 HD-ZIPs |
Manes.03G059200 Manes.04G020100 Manes.04G133100 Manes.11G034600 Manes.11G146000 Manes.14G122900 Manes.16G074600 |
|
miR167 |
3 |
|
Manes.04G102400 Manes.06G167300 Manes.08G092300 |
|
miR168 |
1 |
1 AGO |
Manes.02G219700 |
|
miR169 |
2 |
|
Manes.03G071000 Manes.09G102100 |
|
miR171 |
2 |
2 SCLs |
Manes.02G038400 Manes.05G048700 |
|
miR172 |
17 |
6 AP2s |
Manes.04G027000 Manes.07G101700 Manes.09G080100 Manes.11G139300 Manes.12G106400 Manes.13G120700 |
|
miR319 |
4 |
3 TCPs |
Manes.13G138300 Manes.14G058400 Manes.15G091000 |
|
miR393 |
4 |
2 TIRs; 2 AFBs |
Manes.01G214300 Manes.05G067800 Manes.04G091500 Manes.11G078600 |
|
miR394 |
2 |
2 F-boxes |
Manes.12G010000 Manes.13G010700 |
|
miR395 |
4 |
1 APS |
Manes.15G033600 |
|
miR396 |
20 |
11 GRFs |
Manes.01G041800 Manes.01G264700 Manes.02G031200 Manes.03G039500 Manes.04G144700 Manes.05G183900 Manes.05G190700 Manes.08G160800 Manes.09G129700 Manes.16G096400 Manes.18G049600 |
|
miR399 |
1 |
|
Manes.15G011700 |
|
miR403 |
1 |
|
Manes.01G024400 |
|
miR477 |
3 |
|
Manes.02G097400 Manes.12G139300 Manes.16G020200 |
|
miR530 |
1 |
|
Manes.13G036300 |
|
miR535 |
1 |
|
Manes.16G020000 |
Table 2: List of genes targeted by miRNAs in cassava.
|
miRNA |
Targets ID |
Fold change |
Description |
|
miR156 |
Manes.01G026000 |
-2.86 |
Squamosa promoter-binding-like protein 6 |
|
|
Manes.02G118800 |
5.23 |
Zinc finger domain-containing protein 66 |
|
|
Manes.06G088300 |
4.55 |
DUF863 domain-containing protein |
|
|
Manes.08G081400 |
4.18 |
E3 ubiquitin-protein ligase RDUF1-like |
|
|
Manes.10G052500 |
2.39 |
NAC transcription factors 5 |
|
miR159 |
Manes.11G009900 |
4.76 |
Transcription factor GAMYB-like |
|
miR160 |
Manes.07G099500 |
-2.86 |
Auxin response factor 16 |
|
|
Manes.18G072400 |
2.06 |
Auxin response factor 17 |
|
miR164 |
Manes.06G090500 |
5.68 |
NAC transcription factors 17 |
|
|
Manes.14G080400 |
13.68 |
NAC transcription factors 62 |
|
miR166 |
Manes.04G139200 |
3.08 |
Beta-galactosidase 1 |
|
|
Manes.06G159400 |
2.08 |
Glutamate synthase 1 |
|
|
Manes.18G105400 |
4.56 |
Patellin-3 |
|
miR167 |
Manes.06G167300 |
-2.13 |
Alpha-L-arabinofuranosidase B |
|
miR172 |
Manes.04G054500 |
2.29 |
Transcription factor bHLH128 |
|
|
Manes.07G034600 |
-2.71 |
ABC transporter family protein |
|
|
Manes.09G080100 |
2.26 |
Floral homeotic protein APETALA 2 |
|
|
Manes.S065500 |
2.05 |
B-cell receptor-associated protein 31 |
|
miR319 |
Manes.15G091000 |
-2.38 |
Transcription factor TCP3-like |
|
miR394 |
Manes.13G010700 |
2.19 |
F-box only protein 6 |
|
miR395 |
Manes.12G129400 |
-2.56 |
Probable glucan 1,3-beta-glucosidase A |
|
|
Manes.17G046600 |
3.54 |
VQ motif-containing protein 4 |
|
miR396 |
Manes.01G041800 |
2.03 |
Growth-regulating factor 7 |
|
|
Manes.02G031200 |
5.55 |
Growth-regulating factor 10 |
|
|
Manes.03G039500 |
-2.44 |
Growth-regulating factor 1 |
|
|
Manes.07G044100 |
2.1 |
NBS-LRR resistance protein RGH1 |
|
|
Manes.07G044300 |
4.55 |
NBS-LRR resistance protein RGH2 |
|
|
Manes.07G044700 |
7.27 |
NBS-LRR resistance protein RGH1 |
|
|
Manes.07G045400 |
3.13 |
NBS-LRR resistance protein RGH1 |
|
|
Manes.14G115700 |
-27.1 |
Hypothetical protein |
|
miR477 |
Manes.02G097400 |
2.16 |
Disease resistance RPP13-like protein 4 |
Table 3: Differential expressed miRNA targets in response to cold stress.
Citation: Li S1, Peng M, Cheng Z (2019) Genome-Wide Identification of miRNAs Targets Involved in Cold Response in Cassava. Open Acc J Agri Res: OAJAR-100020.