Emergency Medicine and Trauma Care Journal
Review Article
RNA Interference Activity and Defense Strategy in Insects
Ogunshola OJ1*, Akanbi OM1, Akinyemi IA2, Ajayi OE2 and Qingfa Wu2
1Department of Animal Production and Health, Federal University of Technology Akure, Nigeria
2School of Life Sciences, University of Sciences and Technology of China, China
*Corresponding author: Ogunshola OJ, Department of Animal Production and Health, Federal University of Technology Akure, Ondo State, Nigeria, Tel: +2348032321183; Email: ojogunshola@futa.edu.ng
Citation: Ogunshola OJ, Akanbi OM, Akinyemi IA, Ajayi OE and Qingfa Wu (2019) RNA Interference Activity and Defense Strategy in Insects. Emerg Med Trauma Care J: EMTCJ-100009.
Received date: 24 August, 2019; Accepted date: 17 September, 2109; Published date: 26 September, 2019
Abstract
RNA interference (RNAi) is a powerful tool and a gene silencing mechanism known to knockdown target genes by suppressing the expression of such genes. RNAi has three pathways which are the piwiRNA (piRNA) pathway that performs the defense strategy by defending against the transposable elements; the microRNA (miRNA) and small interfering RNA (siRNA) pathways performing the negative regulation for gene expression. The siRNA pathway is activated by exogenous dsRNA and this pathway serves to defend the genome against invading nucleic acids. RNAi experiments exploit the siRNA pathway by delivering dsRNA to induce knockdown of the target gene. This technology also has many potential applications in molecular biology, agriculture and many other fields. The application of RNAi technology for management of insects, blocking disease transmission by insect vectors and controlling pathogens that affects beneficial insects were reported helpful. Hence, this review showed the activity of RNAi in the knockdown of target genes as defense strategy in insect.
Keywords: Insects; Gene; RNA interference; RNAi; RNA Induced Silencing Complex (RISC)
Introduction
An insect can be defined simply as a small, six-legged animal. More than 800,000 of the million kinds of animals that scientists have described and named are insects [1]. Around 7,000 to 10,000 new kinds of insects are discovered every year and are mostly everywhere from steamy tropical jungles to cold polar regions, from snow-capped mountains to deserts below sea level. Researches has revealed the uses of insects [2,3]. For instance, in agriculture, insects are considered harmful and beneficial. Insects are one of the chief competitors for food and fiber. On yearly basis, insects cause huge damage to field crops, vegetables, fruits and fibers in all stages of growth, production, storage, processing and distribution [4]. However, Insects are not just pests to our society but are still beneficial to humans. Insects serve as source of food and are important part of the food chain because birds and fish eat insects directly to survive. Many mammals and reptiles feed on insects as well. Honey bees do not only make honey but also helps in pollinating plants. Some insects are beneficial in nature as they are helpful to humans by preying on and destroying other insects that are considered harmful. Medicinally, Insects can transmit diseases by many methods and can be used by researchers to find out about diseases. This includes animal and plant diseases as well as human diseases. Insects are used extensively in genetic research because they reproduce efficiently and can be handled so easily in large populations. For these reasons, researchers have been well equipped with a great wealth of knowledge about heredity, biological growth and development, causes and treatments of diseases.
RNA interference (RNAi), a gene-silencing mechanism which provides double stranded RNA molecules matching a specific target gene sequence. RNAi is also known as dsRNA-mediated gene silencing or Post-Transcriptional Gene Silencing (PTGS). This is now widely used in functional genetic studies and its potential application to control agricultural insect pests has rapidly become evident [5].
In the early 1990s, an unexplained genetic phenomenon of gene silencing was observed in Petunia plants. When a gene coding for a key enzyme involved in pigmentation was introduced into the plants to increase production of enzyme resulting in increased conversion of substrate to purple molecules, scientists observed that there was a decrease without any increase in the purple colour [6]. Further studies revealed that aside the introduced gene that was not expressed, there was also a suppression in the endogenous gene coding for this enzyme and this phenomenon was called co-suppression and the exact molecular mechanism was not ascertained [7]. Later on, such gene silencing was reported in viral resistance in plants and animals. When a La Crosse virus gene was inserted into an infectious Sindbis virus expression vector, it caused suppression of virus replication [8]. Similar approaches were also used to engineer resistance to California sero-group virus replication in mosquito cells and mosquitoes [9]. It was also reported in fungus Neurospora crassa, the sequence specific silencing of gene expression but the mechanism of silencing was not fully understood [9]. Functioning of RNAi was demonstrated in nematodes, insects, plants and trypanosomes within three years after the discovery of RNAi [10-16]. In 2001, the first successful RNAi was reported after exposing mammalian cells to the siRNA (Hope, 2001). Scientists had worked on insects e.g. fruit fly, Drosophila melanogaster, with the application of RNAi methods so as to understand the functions of genes coding for proteins involved in their growth, development, reproduction and behavior [17].
RNA interference (RNAi) has enabled the functional analysis of genes in non-model organisms without the requirement of a mutant. In eukaryotes, RNAi is a naturally occurring phenomenon in which a double-stranded RNA suppresses the expression of a target gene. RNAi served as a research tools to entomologist which they used in studying genes that are involved in physiological processes, embryogenesis, reproduction and behavior in model and non-model insects [18].
An emerging area for RNAi research is its use as a species-specific insecticide in insect pest management [19]. RNAi efficacy is highly variable depending upon the insect species [20], the method of RNAi delivery [21] and the gene targeted by RNAi [22]. By gaining a better understanding of the relationship between RNAi delivery and efficacy, as well as identifying the genes that are responsive to RNAi we will better understand mechanisms associated with RNAi in insect systems.
RNAi Pathways and Mechanism
There are three major RNAi pathways found to have been characterized for small non-coding RNAs.These pathways are:
The three RNAi pathways employ several distinct but related core RNAi proteins.The piwiRNA (piRNA) pathway performs the defense strategy by defending against the transposable elements. Both the microRNA (miRNA) and small interfering RNA (siRNA) pathways perform the negative regulation for gene expression [23]. Some studies had made known that all RNAi pathways are evolutionarily conserved and that there is variation in core genes of the small interfering RNA (siRNA) pathway among insects [24,25] examined the conservation of core RNAi proteins between different insect species and C. elegans in the miRNA and siRNA pathways. Core RNAi genes implicated in the miRNA pathway and piRNA pathway are more conserved between species than core RNAi genes of the siRNA pathway.
The siRNA pathway is activated by exogenous dsRNA, and this pathway serves to defend the genome against invading nucleic acids. The dsRNA as the agent responsible for gene silencing phenomenon was discovered in nematode [26]. RNAi experiments exploit the siRNA pathway by delivering dsRNA to induce knockdown of the target gene. When a dsRNA is introduced into a cell, it is processed into ~21bp small interfering RNAs (siRNAs) by an enzyme called Dicer-2. The siRNAs are incorporated into an RNA Induced Silencing Complex (RISC), which is coupled with the argonaute-2 (Ago-2) protein. The double-stranded siRNA is unwound, the passenger strand is degraded and the RISC complex uses the guide strand to guide the complex to the homologous mRNA. The RISC complex has endonuclease activity and cleaves the mRNA. The mRNA is destroyed, and the protein for which the mRNA is coding is not expressed Figure 1 below shows the RNAi pathways and mechanism.
dsRNA Delivery Methods to Insects
In order to induce knockdown of target gene, there must be dsRNA delivery. In insects, different methods of dsRNA delivery are adopted but there arethree most common methods of delivering dsRNAs to insects [27]. These methods are:
Microinjection: Microinjection was one of the first methods used for delivering dsRNAs. dsRNAs was delivered via this method inan insect (Drosophila melanogaster) [28]. This method is used to deliver dsRNAs directly into insect hemolymph or into an insect embryo. This method of delivering dsRNAs has advantages over other delivery methods because the site of delivery and the dose of dsRNAs are controlled.
Feeding: Feeding as a method of delivering dsRNAs to the midgut cells requires the uptake of dsRNAs by the cells. In insect that exhibits a robust RNAi response, this method is confirmed to be effective. In order to avoid the mechanical damage caused by microinjection method of dsRNA delivery in insects, RNAi by feeding can be used to deliver dsRNAs in a high-throughput manner [29]. This method of delivering dsRNA to insect plays an important role in insect control [30].
Soaking: Delivery of dsRNA in insects by soaking method has been mainly conducted in cell lines. According to [31], this method has also been shown to be valuable as a high-throughput tool for large-scale gene expression studies in C. elegans.
Factors to Consider in adopting dsRNA Delivery Method
In choosing methods of dsRNA delivery to insects, the selected methods may be adopted based on:
For instance, dsRNAs are delivered into the midgut or hemolymph for the cell to take up the dsRNAs, and this is mostly done in entomological RNAi experiments that relies on extracellular RNAi [32]. According [33-35] to extracellular RNAi is classified as environmental and/or systemic. The phenomenon in which dsRNAs are taken up from the cells’ environment and the exhibition of gene knockdown effect such cells is referred to as environmental RNAi. Systemic RNAi occurs when the silencing effect is passed from cell to cell [36] while the mechanism for extracellular RNAi is poorly understood. Endocytic pathway and a transmembrane protein, SID-1 are the two proposed mechanisms for the uptake of dsRNAs [37].
The Benefits of RNAi Activity
RNA interference (RNAi) activity is to mediate resistance to both endogenous parasitic and exogenous pathogenic nucleic acids and likewise regulates the expression of protein-coding genes. RNAi suppresses gene expression by small noncoding RNA molecules, mostly by the cleavage of a target mRNA in a sequence-speci?c manner [38].
In plants and animals, RNAi provides one line of defense against RNA viruses and foreign dsRNA molecules. Small endogenous RNAs known as micro RNAs are also processed by a related pathway to regulate tissue-specific patterns of gene expression primarily via translational regulation [39-45]. Long non-coding RNAs also play a prominent role in the epigenetic regulation of gene expression [46]. It is now clear that far more of the genome is transcribed than previously thought [47] and that RNA, in addition to being the obligate messenger and facilitator of protein synthesis in the cell, is also a central player in the regulation of eukaryotic gene expression.
RNAi is used in functional genomics (systematic analysis of loss-of-function phenotypes induced by RNAi triggers) and developing therapies for the treatment of viral infection, dominant disorders, neurological disorders and many types of cancers (in vivo inactivation of gene products linked to human disease progression and pathology). In classical or forward functional genetics, a gene associated with the phenotype is determined [48], usually via a mutant organism. While forward functional genetic practices have proven invaluable for the functional analysis of genes, there are several drawbacks to this method. Forward genetics typically rely on a mutant organism, but a naturally variant or an induced mutant can be difficult to obtain in the laboratory [49].
Additionally, forward functional genetic practices are largely limited to model organisms, such as Drosophila melanogaster or Tribolium castaneum [50]. RNAi enables the application of reverse functional genetics, whereby a gene of interest is chosen and the phenotype associated with the gene is discovered [9]. This has opened the door to our understanding of gene function in non-model organisms. RNAi has advantages over classical functional genomics studies, as the only genetic information required is the DNA sequence of the target gene. Additionally, RNAi allows for a phenotypic evaluation of gene function without the requirement of a mutant organism. RNAi has been used in functional genetics to elucidate genes involved in many insect processes, including embryonic development, physiology and behavior [9]. Embryonic RNAi was utilized to determine the role of two genes, frizzled and frizzled 2, in the Drosophila wingless pathway [51]. Two previously known insect chitin synthase genes were determined to have separate roles by RNAi: CHS1 was found to function solely in the formation of chitin found in the insect cuticle, while CHS2 was found to function mainly in the formation of chitin in the peritrophic membrane [52].
Similarly, RNAi was used in T. castaneum to distinguish the phenoloxidase gene(s) thought to be involved in cuticle tanning. These researchers found that RNAi of the Laccase2 gene caused a loss of pigmentation and sclerotization in the beetle, indicating that Laccase2 was the gene that is required for these biochemical processes. A review of functional RNAi conducted in Tribolium castaneum reveals that RNAi has been used in more than 15 studies to further our understanding of many insect processes.
RNA interference (RNAi) has transformed insect science research because it enables the researcher to suppress a gene of interest and thereby link a phenotype to gene function. For basic research purposes, RNAi offers a route to functional genetics in all insects, including those for which transgene resources do not exist. RNAi also has enormous potential for applied entomology [53,54]. For example, RNAi can be used for insect pest control by suppressing essential genes leading to reduced ?tness and/or mortality. Furthermore, by priming the antiviral RNAi response with innocuous viral sequences, bene?cial insect species, such as honey bee (Apis mellifera) and silkworm (Bombyx mori), can be protected from highly pathogenic viral infections. However, the reality is not yet matching the envisioned potential of RNAi. Practitioners are increasingly aware that RNAi in insects can be capricious; ef?cacy varies across insect taxa, among genes, with mode of delivery, and even between different laboratories [55]. The application of RNAi activity in insects as defense strategy cannot be discarded by researchers due to its benefits.
RNAi as a Defense Strategy in Insects
Pathogens that causes serious diseases attacked many beneficial insects, thereby reducing their ability to perform their beneficial functions. This had led to the discovery of different methods to control pathogens that attacks the beneficial insect. The discovery and development of RNAi and its technology has been adopted to control pathogens that attack beneficial insects. dsRNA was delivered via feeding or injection method to target specific genes in the Israeli acute paralysis virus that affects honey bees. This was done so as to efficiently knockdown the expression of the target genes, thereby reducing the virus disease [56]. In field trials, Remebee-1, which is a dsRNA product from Beeologics Inc., Miami, Florida, USA was successfully tested to control Israeli acute paralysis virus infection of honey bees [57].
RNAi has been used for identifying the target sites for controlling insect pest like red flour beetle (Tribolium castaneum). The dsRNA was delivered via injection method into the haemocoel of the pupae so as to efficiently knockdown the expression of zygotic genes in the embryos developed from the eggs laid by female adults that enclosed from the dsRNA injected pupae. It has also been found that there was a knockdown in the expression of target genes in the pupae as well as adults developed from larvae injected with dsRNA [58]. The injection of minute quantities of dsRNA in Tribolium castaneum showed how highly effective RNAi is, in knocking down gene expression. Also in Diabrotica virgifera virgifera commonly known as western corn rootworm, RNAi has been used to identify target genes that could be used to create a defense strategy for controlling this pest.
In insects like Colorado potato beetle, tephritid fruit fly and beet armyworm, when bacteria expressed dsRNA was fed to these insects led to the efficient induction of RNAi. This method of using bacteria to deliver dsRNA to insect, is a possible method or defense strategy that could be used for pests’ management. The cost per application is an important factor in choosing dsRNA for pest management and using bacterially expressed dsRNA is of lower cost when compared with in vitro synthesized dsRNA. Bacterially expressed dsRNA could be sprayed on the crop plants whenever necessary due to the lower costs of application and the ease in manufacturing large quantities. In several RNAi studies, gene expression is not knockout but rather knockdown, therefore rendering such genes and their effects silence. The delivery of dsRNA for the target gene is done continuously so as to avoid the risk of resistance development. This has made the dsRNA-based insecticides to have advantages when compared with the chemical insecticides in term of overcoming insecticide resistance.
In understanding host parasite interactions in insects that transmit pathogens causing deadly humans diseases with a view of using this information to set a defense strategy that could block the transmission of pathogens by insects, RNAi methods are used. dsRNA has been delivered into Anopheles gambiae adults via injection method to cause an effective silencing of genes, which plays an important role in mosquito antimicrobial defence against Gram-positive bacteria [58]. Injecting dsRNA in Anopheles gambiae was used to determine the function of a complement-like protein, TEP1. This led to a knockdown in the TEP1 gene expression which increased the number of developing parasites in the susceptible strain. The injection of dsRNA into adults to determine the function of various genes involved in host-parasite interactions and immune response has been seen in several studies using different mosquito species. One of the best methods used to produce transgenic insects that are refractive is the RNAi, as several studies had shown that a method to prevent transmission of diseases by insect vectors is via the use of genetically transgenic insects that are refractive to diseases-carrying pathogens to replace natural populations of insect vectors.
Conclusion
This piece of review indicated that RNAi is a successful methodthat could be used to improve knockdown efficiency of target genes in insects, hence creating a defense strategy for beneficial insects against parasites and pathogens. It is possible to feed dsRNA and achieve silencing of target genes in several insect species. However, the cost-effective methods of producing dsRNA in large quantities is the major challenge in achieving insect control by feeding. There is need for more researches to be done to improve knockdown efficiency of target genes in sucking insects, development of dsRNA delivery methods and efficient production of dsRNA. This will help manage insect pests, disease carrying vectors and pathogens causing diseases in beneficial insects.
Authors’ contributions
OOJ wrote the manuscript. QW was the principal author who was responsible to manage all activities of the review. OOJ, AOM, AIA and AOE read the second draft of the manuscript. All authors read and approved the final manuscript.
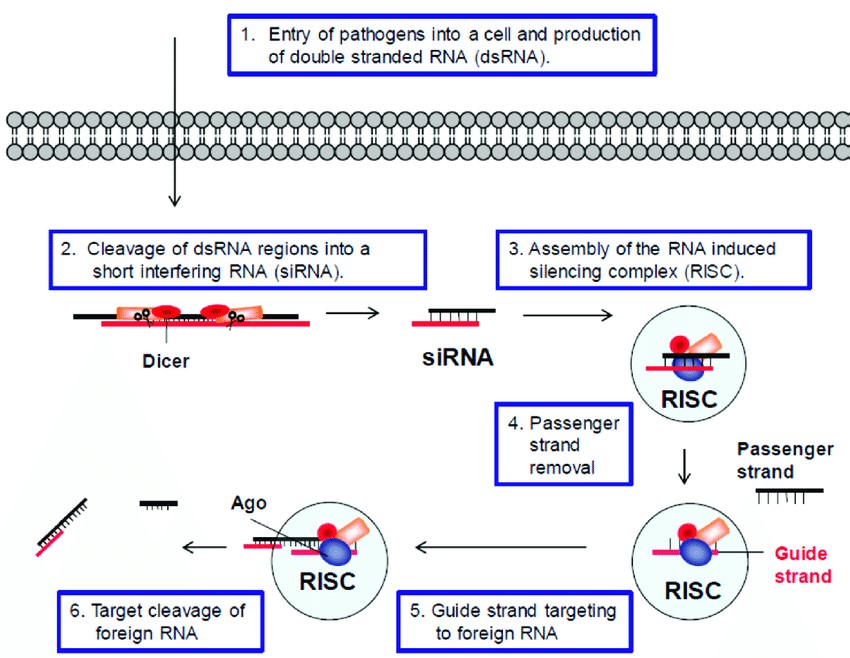
Figure 1: RNAi defense pathway.
Citation: Ogunshola OJ, Akanbi OM, Akinyemi IA, Ajayi OE and Qingfa Wu (2019) RNA Interference Activity and Defense Strategy in Insects. Emerg Med Trauma Care J: EMTCJ-100009.